
Dr Tamas Korcsmaros recently joined MDR as a Senior Lecturer in Intestinal Epithelial Biology, in the Section of Hepatology and Gastroenterology. We caught up with him to find out about his background, research and interests.
Introduce yourself – who are you and what do you do?
I am Tamas Korcsmaros, originally from Hungary, and I have been living in the UK since 2014. I am a systems biologist working with both computational and experimental approaches to study signalling networks in the gut. I am particularly interested in how cell-cell and cell-microbe interactions affect intestinal homeostasis, and how we could use precision medicine to tackle current challenges to treat patients with inflammatory bowel disease.
Where were you working before you joined MDR?
I was originally trained as a biochemist, then I did a PhD in computational and systems biology in the Semmelweis University (the medical university in Budapest, Hungary), which was followed by establishing a small bioinformatics group at the Department of Genetics to develop computational resources for the community. This is where I started doing research on autophagy, a key cellular process for all our cells. Then in 2014, I got a unique fellowship to establish my group in Norwich. Here, I had a joint appointment between two BBSRC funded institutes, the sequencing and computational biology focused Earlham Institute and the gut health and microbiome focused Quadram Institute. When I now left Norwich, I was a Tenure-track group leader and the lead on the Earlham Institute’s strategic programme on systems genomics.
What first inspired your interest in intestinal epithelial biology?
The intestinal epithelium is a single cell layer dividing our body from the gut luminal content, including nutrients and microbes. It is basically the first line of defensein our gut. It also regulates both the microbiota composition and the regeneration and inflammatory processes of our cells. I found it fascinating how specific cells in our gut epithelium can do these things, and how they work together with commensal bacteria to achieve their normal function.
What is the most interesting piece of research you are currently working on?
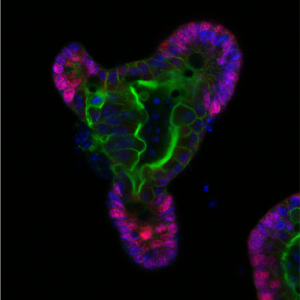 The intestinal epithelium unfortunately often cannot or does not work properly. This is the situation for example in inflammatory bowel disease (IBD), a chronic pathology of the gut that affects around 7 million people worldwide (from which 500,000 patients are in the UK) and costs the NHS >£1.8 billion annually. I am interested to find ways to improve how the epithelial layer works in patients with IBD, and how we can best match the efficient treatments with those patients who would respond to them. I am investigating this challenging and exciting question with computational modeling (predicting metabolite and microbial connections to epithelial cells in healthy and in diseased states) and with experimentally testing the gut epithelial interface with a very versatile and useful model system, called organoids. Since its first publication in 2009, the in vitro gut organoid model has been recognized as a major technological breakthrough tool in many basic biology and clinical applications. Gut organoids are near-physiological 3D model systems that facilitate studying a range of in vivo biological processes including cell differentiation, anti-microbial peptide production, host-microbe interactions and cell-cell communications. Organoids can also be used to examine the effect of defined mutations by generating transgenic organoids. They are a powerful and promising tool to investigate the physiological and cell-specific effects of disease-related mutations, especially in IBD.
The intestinal epithelium unfortunately often cannot or does not work properly. This is the situation for example in inflammatory bowel disease (IBD), a chronic pathology of the gut that affects around 7 million people worldwide (from which 500,000 patients are in the UK) and costs the NHS >£1.8 billion annually. I am interested to find ways to improve how the epithelial layer works in patients with IBD, and how we can best match the efficient treatments with those patients who would respond to them. I am investigating this challenging and exciting question with computational modeling (predicting metabolite and microbial connections to epithelial cells in healthy and in diseased states) and with experimentally testing the gut epithelial interface with a very versatile and useful model system, called organoids. Since its first publication in 2009, the in vitro gut organoid model has been recognized as a major technological breakthrough tool in many basic biology and clinical applications. Gut organoids are near-physiological 3D model systems that facilitate studying a range of in vivo biological processes including cell differentiation, anti-microbial peptide production, host-microbe interactions and cell-cell communications. Organoids can also be used to examine the effect of defined mutations by generating transgenic organoids. They are a powerful and promising tool to investigate the physiological and cell-specific effects of disease-related mutations, especially in IBD.
Do you have any collaborative opportunities in your research?
I am a very collaborative person. The computational tools we develop are always shared with the community with tutorials and open-source codes. Still, we are happy to provide support to adapt and use these resources for colleagues interested in using them for their studies in signalling pathways, autophagy, antioxidant response, and host-microbe interaction projects. At Imperial, I am looking forward to working together with bioengineers, cell biologists, immunologists, and microbiologists as well as omics and clinical researchers to establish new organoid-based models that could be used to improve our understanding on biological and medical processes.
What has been the proudest achievement of your career so far?

During my PhD, I created a new signaling network resource, called SignaLink. In the last 15 years, we have continuously improved and upgraded this gap-filling resource, and now it has become one of the gold standard signaling resources that has been used by more than 400 institutes in 81 countries, and we recorded at least 25,000 visits to this database. In the last 6-8 years, we have been using SignaLink to understand better which genes are mutated and which genes are differentially expressed in diseased states (eg in different types of cancer or IBD). We described a fascinating general rule, independent of the disease type or tissue that seems to govern how pathologic rewiring is happening.
What do you find to be the most challenging aspect of your work?
I am a multi-disciplinary researcher, and I have always been inspired to work with people with different backgrounds and expertise. I am always motivated to bridge people with relevant expertise to tackle complex questions. This, however, requires a general understanding of different fields, learning their terminologies, culture and understanding their motivation to engage in collaborative projects. It is a fruitful and exciting thing to do – but often challenging as it needs more time, and you could easily become a ‘Jack of all trades’ if you don’t keep a well-defined focus.
What do you consider to be the key challenges in healthcare and medicine that we need to overcome in the next 20 years?
Precision medicine and personalised microbial therapies. They are connected and both hold great promise. While there will be new drugs developed that will be better than the current ones, precision medicine has the promise to exploit the existing drugs better, by giving them to patients who have a high chance of responding. Modulating our health with microbes is possible and potentially less dramatic than pharmacological treatments but, currently, we do not have enough data to understand which strain is working, and when or why. An extra challenge is that patients are often different, and their medical history on what antibiotics treatment they received earlier, or even the mode of how they were born could affect the efficacy of any microbial supplementation. In the next 20 years, we need to learn more about these topics and about how we can support the ‘good bugs’ in our gut with an improved and healthy diet.
What is the best piece of advice you would give to students and early career researchers who aspire to have a successful career in research and/or academia?
Get used to living with failures, learn from them, move on, and enjoy every success you achieve together with all your colleagues and friends who helped and supported you.
And finally, if you could time travel, when/where would you go?

Probably it would be sometimes in the late medieval ages and the Renaissance period. It would be nice to travel to some of the main European cities at that time, see how they were constructing the by now classical buildings, talking to the inventors and nature researchers about their view of our body and physical world.