
PhD student Philippa May reflects on being a scientist in the field blood cancer, from working in a leukaemia diagnostic laboratory to a research laboratory.
For the last 10 years I have been a clinical scientist in genetics working across various London NHS Trusts. Whilst I loved diagnostics, last year I left my job to complete my PhD. I worked in a part of life sciences called cytogenetics. This meant when a patient was diagnosed with blood cancer, I would analyse their chromosomes – the structures into which DNA is organised – from their blood or bone marrow to look for specific abnormalities. For some patients, this can lead to a definitive diagnosis. For others a refined prognosis, and in some, it’s simply a way of monitoring how well the patient’s leukaemia is responding to their treatment.
Blood cancer can be very straightforward to diagnose and it was perfectly possible to provide genetic confirmation of a blood cancer diagnosis in a matter of hours. For example, in patients with chronic myeloid leukaemia (CML), I would find a particular abnormality called a Philadelphia translocation between chromosomes 9 and 22. Finding this translocation means a patient will benefit from a targeted therapy – called a tyrosine kinase inhibitor (TKI) – which reverses the effect of the translocation with relatively few side effects. TKIs are a tablet taken once or twice a day at home. Compared to chemotherapy, TKIs have revolutionised the treatment and outcomes of CML, which has been life-changing for CML patients. It was always satisfying to call the referring clinician and let them know their patient had a Philadelphia translocation because I knew that would set the wheels in motion for a TKI to be prescribed. Ultimately I knew I had made a difference to a patient on those days.
The lab had little direct patient contact, but the nature of the work meant that we had ‘our regulars’; the patients who needed frequent monitoring following a treatment change, or a bone marrow transplant, for example. I have a terrible memory for names/faces in everyday life, but for some reason, I retain names/chromosomal abnormalities very readily. I would always recognise a returning patient’s name and know what abnormality we were hoping not to see again.
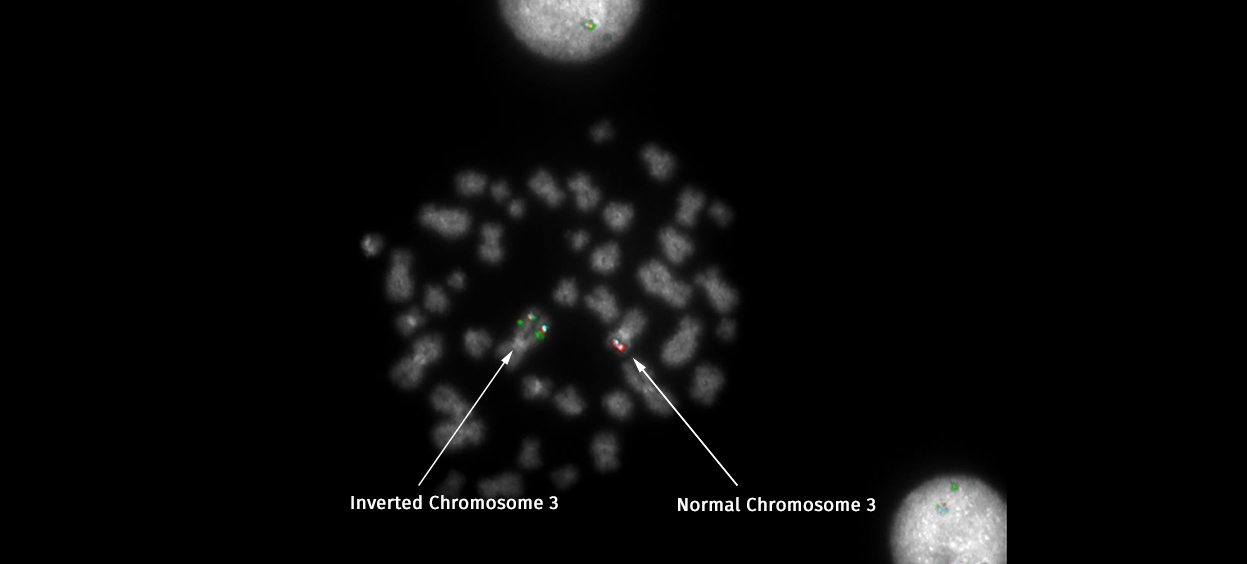
Of course, the nature of working in a leukaemia laboratory meant that there was often bad news to deliver too. At times like that, I could only strive to deliver it as rapidly as possible to help the clinician to decide what treatment was appropriate. One abnormality which falls into the bad news category is a rearrangement of the long arm of chromosome 3 which switches on an oncogene – a gene that can cause cancer – called EVI1. A rearrangement of this gene has one of the most universally poor prognoses and was always news I delivered with a heavy heart. With no targeted therapy for this rearrangement, this news meant – if the patient was well enough – they would undergo a grueling regime of intense chemotherapy and possibly a bone marrow transplant.
It is no surprise then, I suppose, that I find myself right at the start of my new, alternative career in research with that very gene on the top of my list of things to investigate: EVI1. Despite having such a hideous prognosis, we still don’t understand why the gene is switched on in many cases. If we could understand why it was switched on, we might be able to develop a drug to switch it back off again, just as we now have for the Philadelphia translocation. So perhaps working in diagnostics had a more direct, more immediate link to the patients; but now, working in research, I feel like I can do more than just deliver the bad news. Maybe one day I can do something about it.
One of the most difficult things I have had to adjust to in research is the failure rate. In diagnostics, failure is unacceptable. We had well-optimised protocols in place that we couldn’t deviate from without very careful validation. On the odd occasion we couldn’t get a result it was usually because of something out of our hands; the patient might have a very low white cell count or the biopsy taken might be too small. In research, however, I am constantly trying out new techniques and new approaches; it’s disheartening when it doesn’t work but it’s soon forgotten when you finally crack it and get an answer.
I’m often asked by colleagues: which career path is more enjoyable, more rewarding, more stressful? It’s very difficult to answer. I enjoy both hugely; I think a happy research lab and a happy diagnostic lab have fundamentally far more in common than the differences between them. There are administrative differences, of course – notably job security (I gave up a permanent contract which was uncharacteristically reckless), salary, the requirement for professional registration and the availability and accessibility of the formal training – but essentially researchers and diagnosticians are all working to try to make things better for patients with blood cancer.
My ultimate aim is to pursue a clinical/academic career where I can continue both diagnostics and research in the same field. I want to finish by extending a thank you to all the patients who consent to have samples taken for research; I don’t often get a chance to say it directly but it really is important for researchers to have access to samples.
Philippa May (@IamPhilippaMay) is a final year PhD student in the Feldhahn Group (Department of Medicine), under the supervision of Dr Niklas Feldhahn and Dr Alistair Reid. Philippa is also an HCPC registered Clinical Scientist in Genetics and has spent the last eight years working in leukaemia diagnostic genetics.
The Feldhahn Group is supported by a Bennett Fellowship from Bloodwise.
Following the launch of the Faculty of Medicine’s reorganised academic structure on 1 August 2019, this post was recategorised to Department of Immunology and Inflammation.